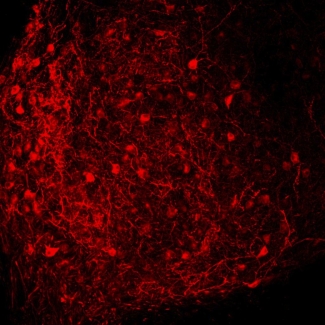
Directing evolution is challenging. The first step requires creating a library of hundreds of thousands of cells containing different mutations of a single protein. This step is now relatively easy, thanks to the tools of molecular biology. The second step requires screening the fluorescence properties of each cell to select only those few that contain top-performing mutant proteins.
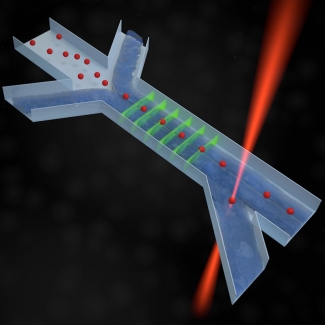
To accomplish the selection process, the group uses microfluidics combined with several laser beams. Its microfluidics system contains micron-sized three-dimensional transparent channels that carry small streams of liquid and allow cells to flow through them one at a time. As the mutant cells pass through the microfluidics channel, lasers measure the fluorescent properties of each mutant cell to assess how well the cells maintain their fluorescence when repeatedly excited by the series of laser beams. Another laser acts as an optical trap that works like a tractor beam to grab onto the best mutant cells for further investigation. The microfluidics setup itself readily removes the cells that are poor performers by simply allowing them flow out of the device.
To make matters more challenging, directed evolution requires repeating the two steps described above multiple times. The Jimenez group is currently in the middle of round three of its quest to evolve a better red-fluorescent protein.
Although the group has already shown that the specific improvements suggested by the computer simulation don’t work, the first round of the directed evolution experiment has come up with an improved red-fluorescent protein with a less floppy barrel that is 2–4 times more stable than mCherry. The combination of mutations that resulted in this improvement has not been previously observed in nature and was completely unexpected.
The group named its new mutant protein Kriek, after a Belgian beer made via the fermentation of cherries. Clearly, the researchers are adept at doing more than biophysics. They include JILA Ph.D. Jennifer Lubbeck (2013) and Fellow Ralph Jimenez, Kevin Dean and Amy Palmer of CU’s Department of Chemistry and Biochemistry, as well as colleagues from the University of Tennessee Space Institute and Florida International University.–Julie Phillips